Mike's fancy plots (xfig example)
--- //example added by Manfred// ---
Mike's fancy plots
A passionate ISIS user (hereafter referred to as "Mike") has compiled a set of powerful plot functions
for spectra in ISIS, which rely on its PGPLOT module. To my knowledge, these functions do currently (2011 April) not yet offer an interface
to extract the data directly within ISIS. However, there is a write_plot function that "creates ASCII files with the data from the last plot made using plot_counts, plot_data, plot_residuals, or plot_unfold." The numbers that appear in any of these fancy plots can hence be dumped to a file, subsequently read from S-Lang, and plotted using SLxfig.
Ideally, one should at some point modify Mike's fancy plot functions, creating a public interface for the data. But for the time being, using write_plot is a simple workaround, and it's actually quite useful to have the data, which shall be displayed in your plots, stored in plain ASCII files -- independent of the full set of spectra and response matrices etc.
If get an error message
fpd is undefined
(or the like) from write_plot, you are most probably using an old version of Mike's scripts (0.X), which is not supported any more, and should therefore seriously consider to change to version 1.x -- even though it is not 100% backward compatible.
An example
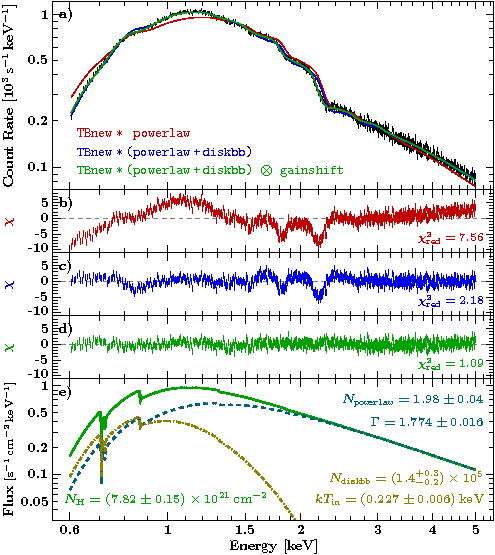
This example is based on Fig. 2.79 of a document I was preparing recently...
As usually, it is recommended to store all files needed to generate //a single plot// for your thesis, talk, publication, etc. in //a single subdirectory//. This advice also holds for the script that loads the data:
Loading the data
You certainly have already a script that loads your data, don't you? It's nevertheless reasonable to copy this file to the directory with the files needed for the plot, since you might modify the load data script in your analysis script, but will once have to find out what (the heck) you did (a long time before) to create (exactly) this plot.
% loaddata.sl
require("isisscripts");
variable dir = getcwd(); % store current working directory
()=chdir("/path/to/your/data"); % go to your analysis directory
variable pn = load_data("src_sd.pha"); % load the data (obviously)
()=chdir(dir); % go back to the previous directory
group(pn; min_sn=20, bounds=0.6, unit="keV"); % do all the setup...
ignore_en(pn, , 0.6);
ignore_en(pn, 5, );
Even though loaddata.sl is here only needed to get the data (next section), it might be useful to keep it separate.
Getting the data / model / residuals
I want to compare three models, for which I copied the parameter files powerlaw.par, powerlaw_diskbb.par, and powerlaw_diskbb_gainshift.par to the directory storing all information how the plot was created.
()=evalfile("loaddata.sl");
variable m;
foreach m (["powerlaw"+["", "_diskbb"+["", "_gainshift"]]]) % a Horner scheme for file names? ;-)
{
message(m);
load_par(m+".par"); % load the model
()=eval_counts(); % evaluate the model
plot_data(1; res=1); % call Mike's fancy plot function <==
write_plot(m); % dump data into ASCII file <==
}
The statements marked by <== create the ASCII data files
powerlaw_0.dat, powerlaw_diskbb_0.dat, and powerlaw_diskbb_gainshift_0.dat
in the following format
# Plot Type : plot_data # # X-axis : \frEnergy (keV) # Y-Axis : \frCounts s\u-1\d keV\u-1\d # Residual : \fr\gx # # Index- 1, Model Plotted- yes, Data File: src_sd.pha/part=0/order=0, Scale: 1.0000e+00 # # Background included in data and model- no # # XAXIS_COLS : 1 set (X 2 columns - bin_lo, bin_hi) # DATA_COLS : 1 set (x 2 columns - Data, Error) # MODEL_COLS : 1 set (x 1 column - Value) # RESIDUAL_COLS: 1 set (x 3 columns - Mean, Mean-1Sigma, Mean+1Sigma)
| X_LO | X_HI | DATA_VALUE | DATA_ERROR | MODEL_VALUE | RES_VALUE | RES_VAL-ERR | RES_VAL+ERR |
|---|---|---|---|---|---|---|---|
| ... | ... | ... | ... | ... | ... | ... | ... |
Reading and plotting the data
Now I can go on with these ASCII data files only, even on my own laptop.
Some global definitions
require("isisscripts");
variable file = ["powerlaw"+["", "_diskbb"+["", "_gainshift"]]];
variable chi2red = [`7.56`, `2.18`, `1.09`];
variable W=14, H=6, % width and height of first panel
Emin=.55, Emax=5.5, % common xrange of all panels
Rmin=-11, Rmax=9, % common yrange of the residual panels
Maj=[.6, 1, 1.5, 2, 3, 4, 5], % major tic marks (energy axis)
Min=[[.5:2:.1], [2:6:.2]]; % minor tic marks (energy axis)
xfig_new_color("col0", 0xC00000);
xfig_new_color("col1", 0x0000E0);
xfig_new_color("col2", 0x00A000);
Data panel
edit: alternatively to the example below using readcol, you can also use
Structure str = read_data_from_write_plot(String_Type file)
...with readcol:
% reading four columns of the first ASCII data file (X_LO, X_HI, DATA_VALUE, DATA_ERROR)
variable lo, hi, val, err; (lo, hi, val, err) = readcol(file[0]+"_0.dat", 1, 2, 3, 4);
variable X = xfig_plot_new(W, H);
X.world(Emin, Emax, min_max(val/1e3; logpad=0.05); xlog, ylog);
X.xaxis(; major=Maj, minor=Min, ticlabels2=0);
X.ylabel(`Count Rate [$10^3\rm\,s^{-1}\,keV^{-1}$]`);
% plotting the data
X.plot((lo+hi)/2., val/1e3, err/1e3; sym="point", size=-1, eb_factor=0, depth=100);
% I add some labels using the world0 coordinate system (in which the plot window extends from 0 to 1).
% I use `` as string delimiters for LaTeX code and avoid to have to escape every backslash (as in "\\").
X.xylabel(.05, .3, `\tt TBnew\:*\: powerlaw`, -.5, 0; world0, color="col0");
X.xylabel(.05, .2, `\tt TBnew\:*\:(powerlaw\,+\,diskbb)`, -.5, 0; world0, color="col1");
X.xylabel(.05, .1, `\tt TBnew\:*\:(powerlaw\,+\,diskbb) $\otimes$ gainshift`, -.5, 0; world0, color="col2");
X.xylabel(.01, 1, `a)`, -.5, .8; world0);
Loop over the models, creating the panels with the residuals
variable R = Struct_Type[3];
variable i;
_for i (2, 0, -1)
{
% read (MODEL_VALUE, RES_VALUE) from the corresponding ASCII data file:
variable m, r; (m, r) = readcol(file[i]+"_0.dat", 5, 6);
% add model to the data panel
X.plot((lo+hi)/2., m/1e3; color="col$i"$);
% create residual panel
R[i] = xfig_plot_new(W, H/3);
R[i].world(Emin, Emax, Rmin, Rmax; xlog);
R[i].yaxis(; major=[-10:10:5], minor=[-20:10], ticlabels2=0);
R[i].xaxis(; major=Maj, minor=Min);
R[i].ylabel(`$\chi$`; color="col$i"$);
R[i].plot((lo+hi)/2., r, 1; sym="point", size=-1, eb_factor=0, color="col$i"$);
R[i].plot([0,1], [0,0]; line=1, color="#808080", depth=100, world01);
R[i].xylabel(.98, 0, `\footnotesize$\chi^2_{\rm red}=`+chi2red[i]+"$", .5, -1; world0, color="col$i"$);
R[i].xylabel(.01, 1, sprintf("%c)", 'b'+i), -.5, .8; world0);
}
Panel with the flux model and its components
variable M = xfig_plot_new(W, H*.75);
M.world(Emin, Emax, 0.03, 1.2; xlog, ylog);
M.xaxis(; major=Maj, minor=Min, ticlabels2=0);
M.xlabel(`Energy [keV]`);
M.ylabel(`Flux \footnotesize[s$^{-1}$\,cm$^{-2}$\,keV$^{-1}$]`);
M.xylabel(.01, 1, `e)`, -.5, .8; world0);
% evaluate the model on a fine grid
load_par(file[-1]+".par");
variable Lo, Hi; (Lo, Hi) = log_grid(.6, 5, 1000);
variable y = eval_fun_keV(Lo, Hi)/(Hi-Lo); % flux in photons/s/cm^2/keV
variable e = sqrt(Lo*Hi); % Since the grid is logarithmic, I tend to use the geometric mean energy.
M.plot(e, y; xlog, ylog, color="col2", depth=90, width=3);
% disk flux
variable coldisk = "#908000";
variable ff = get_fit_fun();
fit_fun(strreplace(ff, "powerlaw(1)+", ""));
y = eval_fun_keV(Lo, Hi)/(Hi-Lo);
M.plot(e, y; color=coldisk, line=3, width=3);
% powerlaw flux
variable colpow = "#006080";
fit_fun("powerlaw(1)*tbnew_simple(1)");
y = eval_fun_keV(Lo, Hi)/(Hi-Lo);
M.plot(e, y; color=colpow, line=1, width=3);
% some labels
M.xylabel(.98, .85, `\small$N_{\tt powerlaw} = 1.98\pm0.04$`, .5, 0; world0, color=colpow);
M.xylabel(.98, .7, `\small$\Gamma = 1.774\pm0.016$`, .5, 0; world0, color=colpow);
M.xylabel(.98, .3, `\small$N_{\tt diskbb} = (1.4^{+0.3}_{-0.2})\times10^{5}$`, .5, 0; world0, color=coldisk);
M.xylabel(.98, .15, `\small$kT_{\rm in} = (0.227\pm0.006)\rm\,keV$`, .5, 0; world0, color=coldisk);
M.xylabel(.02, .15, `\small$N_{\rm H}=(7.82\pm0.15)\times10^{21}\rm\,cm^{-2}$`, -.5, 0; world0, color="col2");
Finally combining all panels
simply using xfig_multiplot:
xfig_multiplot(X, R, M; xlabel=`Energy [keV]`).render("EPIC-pn_spectrum.pdf");
The result
Conclusion: An EPIC-pn burst mode spectrum may require a gainshift correction!