Difference between revisions of "Slurm"
| Line 28: | Line 28: | ||
==== Get cluster status ==== | ==== Get cluster status ==== | ||
In order to get an overview of the cluster, type | In order to get an overview of the cluster, type | ||
| − | + | <pre> | |
sinfo | sinfo | ||
| − | + | </pre> | |
This command offers a variety of options how to format the output. In order to get a detailed output while focusing on the nodes rather than the partitions, type | This command offers a variety of options how to format the output. In order to get a detailed output while focusing on the nodes rather than the partitions, type | ||
| − | + | <pre> | |
sinfo -N -l | sinfo -N -l | ||
| − | + | </pre> | |
An overview over the available partitions can be shown with | An overview over the available partitions can be shown with | ||
| − | + | <pre> | |
scontrol show partition | scontrol show partition | ||
| − | + | </pre> | |
an the current queued and running jobs can be displayed using | an the current queued and running jobs can be displayed using | ||
| − | + | <pre> | |
squeue | squeue | ||
| − | + | </pre> | |
| Line 48: | Line 48: | ||
In order to allocate for instance 1 CPUs and 100MB of memory for real-time work, type | In order to allocate for instance 1 CPUs and 100MB of memory for real-time work, type | ||
| − | + | <pre> | |
salloc --mem-per-cpu=100 -n1 bash | salloc --mem-per-cpu=100 -n1 bash | ||
| − | + | </pre> | |
Your bash is now connected to the compute nodes. In order to execute a script use the srun-command | Your bash is now connected to the compute nodes. In order to execute a script use the srun-command | ||
| − | + | <pre> | |
srun my_script.sh | srun my_script.sh | ||
| − | + | </pre> | |
Each srun-command you execute now is interpreted as a job step. The currently running job step of submitted jobs can be displayed using | Each srun-command you execute now is interpreted as a job step. The currently running job step of submitted jobs can be displayed using | ||
| − | + | <pre> | |
squeue -s | squeue -s | ||
| − | + | </pre> | |
You can also start a job by simply use the srun command and specify your requirements. In the following case, srun will allocate 100MB of memory and 1 CPU(s) for 1 task, only for the duration of execution. | You can also start a job by simply use the srun command and specify your requirements. In the following case, srun will allocate 100MB of memory and 1 CPU(s) for 1 task, only for the duration of execution. | ||
| − | + | <pre> | |
srun --mem-per-cpu=100 -n1 my_script.sh | srun --mem-per-cpu=100 -n1 my_script.sh | ||
| − | + | </pre> | |
If resources are available your job will start immediately. | If resources are available your job will start immediately. | ||
| Line 77: | Line 77: | ||
The ''srun'' commands in the job script launch the job steps. The example below thus consists of two job steps. Each of the ''srun'' commands may have own requirements concerning memory any may also spawn less tasks than given in the header of the script file. However, the values in the header may never be exceeded! | The ''srun'' commands in the job script launch the job steps. The example below thus consists of two job steps. Each of the ''srun'' commands may have own requirements concerning memory any may also spawn less tasks than given in the header of the script file. However, the values in the header may never be exceeded! | ||
| − | + | <pre> | |
#!/bin/bash | #!/bin/bash | ||
#SBATCH --job-name my_first_job | #SBATCH --job-name my_first_job | ||
| Line 88: | Line 88: | ||
srun -l my_script1.sh | srun -l my_script1.sh | ||
srun -l my_script2.sh | srun -l my_script2.sh | ||
| − | + | </pre> | |
The ''-l'' parameter of ''srun'' will print the task number in front of a line of stdout/err. You can submit this script by saving it in a file, e.g. ''my_first_job.slurm'', and sumitting it using | The ''-l'' parameter of ''srun'' will print the task number in front of a line of stdout/err. You can submit this script by saving it in a file, e.g. ''my_first_job.slurm'', and sumitting it using | ||
| − | + | <pre> | |
cepheus:~> sbatch my_first_job.slurm | cepheus:~> sbatch my_first_job.slurm | ||
Submitted batch job 144 | Submitted batch job 144 | ||
| − | + | </pre> | |
You can check the estimated starting time of your job using | You can check the estimated starting time of your job using | ||
| − | + | <pre> | |
squeue --start | squeue --start | ||
| − | + | </pre> | |
==== Submitting a job array ==== | ==== Submitting a job array ==== | ||
| Line 104: | Line 104: | ||
In order to submit a array of jobs with the same requirements you have to modify your script file. | In order to submit a array of jobs with the same requirements you have to modify your script file. | ||
The script above is going to spawn 4 jobs each consisting of one srun command. Note the presence of the new environment variable ''${SLURM_ARRAY_TASK_ID}'' which might be useful for your work. In this example we start an isis-script with different input values. You can also simply use different scripts. | The script above is going to spawn 4 jobs each consisting of one srun command. Note the presence of the new environment variable ''${SLURM_ARRAY_TASK_ID}'' which might be useful for your work. In this example we start an isis-script with different input values. You can also simply use different scripts. | ||
| − | + | <pre> | |
#!/bin/bash | #!/bin/bash | ||
#SBATCH --partition remeis | #SBATCH --partition remeis | ||
| Line 122: | Line 122: | ||
srun /usr/bin/nice -n +19 ${COMMAND[$SLURM_ARRAY_TASK_ID]} | srun /usr/bin/nice -n +19 ${COMMAND[$SLURM_ARRAY_TASK_ID]} | ||
| − | + | </pre> | |
As above, this code might be saved in a file, for example ''job.slurm'' can be executed using | As above, this code might be saved in a file, for example ''job.slurm'' can be executed using | ||
| − | + | <pre> | |
sbatch job.slurm | sbatch job.slurm | ||
| − | + | </pre> | |
If you need a specific machine to run your job on, you can use | If you need a specific machine to run your job on, you can use | ||
| − | + | <pre> | |
#SBATCH --nodelist=leo,draco | #SBATCH --nodelist=leo,draco | ||
| − | + | </pre> | |
If you have a job with high I/O and/or traffic on the network you can limit the number of jobs running simultaneously (to 2 in this example) by | If you have a job with high I/O and/or traffic on the network you can limit the number of jobs running simultaneously (to 2 in this example) by | ||
| − | + | <pre> | |
#SBATCH --array 0-3%2 | #SBATCH --array 0-3%2 | ||
| − | + | </pre> | |
SLURM will only allocate resources on the given nodes. However, if all nodes in 'nodelist' cannot fulfill the job requirements, SLURM will also allocate other machines. | SLURM will only allocate resources on the given nodes. However, if all nodes in 'nodelist' cannot fulfill the job requirements, SLURM will also allocate other machines. | ||
If you would like to cancel jobs 1, 2 and 3 from job array 20 use | If you would like to cancel jobs 1, 2 and 3 from job array 20 use | ||
| − | + | <pre> | |
scancel 20_[1-3] | scancel 20_[1-3] | ||
| − | + | </pre> | |
If you want to cancel the whole array, ''scancel'' works as usual | If you want to cancel the whole array, ''scancel'' works as usual | ||
| − | + | <pre> | |
scancel 20 | scancel 20 | ||
| − | + | </pre> | |
Note, there is also the option to modify requirements of single jobs later using ''scontrol update job=101_1 ...''. | Note, there is also the option to modify requirements of single jobs later using ''scontrol update job=101_1 ...''. | ||
| Line 153: | Line 153: | ||
In order to allow each command of the job array to change into an individual directory (as opposed to all into the same directory as above), modify the script as follows: | In order to allow each command of the job array to change into an individual directory (as opposed to all into the same directory as above), modify the script as follows: | ||
| − | + | <pre> | |
#!/bin/bash | #!/bin/bash | ||
#SBATCH --partition remeis | #SBATCH --partition remeis | ||
| Line 176: | Line 176: | ||
srun /usr/bin/nice -n +19 ${COMMAND[$SLURM_ARRAY_TASK_ID]} | srun /usr/bin/nice -n +19 ${COMMAND[$SLURM_ARRAY_TASK_ID]} | ||
| − | + | </pre> | |
This also works with paths relative to the directory where the slurm script was submitted. | This also works with paths relative to the directory where the slurm script was submitted. | ||
==== Using the ''erosita'' partition (serpens)==== | ==== Using the ''erosita'' partition (serpens)==== | ||
If you are allowed to use the eRosita Partition, contact a SLURM admin (eg. [[mailto:simon.kreuzer@fau.de|simon.kreuzer@fau.de]]). Once your username is added to the list of privileged users, you just have to add | If you are allowed to use the eRosita Partition, contact a SLURM admin (eg. [[mailto:simon.kreuzer@fau.de|simon.kreuzer@fau.de]]). Once your username is added to the list of privileged users, you just have to add | ||
| − | + | <pre> | |
#SBATCH --partition=erosita | #SBATCH --partition=erosita | ||
#SBATCH --account=erosita | #SBATCH --account=erosita | ||
| − | + | </pre> | |
to your jobfiles. | to your jobfiles. | ||
| Line 209: | Line 209: | ||
MPI (the Message Passing Interface) makes it possible to run parallel processes on CPUs of different hosts. To do so it uses TCP packets to communicate via the normal network connection. Some tasks can profit a lot of using more cores for computation. | MPI (the Message Passing Interface) makes it possible to run parallel processes on CPUs of different hosts. To do so it uses TCP packets to communicate via the normal network connection. Some tasks can profit a lot of using more cores for computation. | ||
At Remeis MPICH2 is used for initialisation of MPI tasks which is well supported within Slurm. The process manager is called **pmi2** and is set as default for srun. If an older MPI process manager is needed, for example for older MPI applications used in **torque**, it can be set with | At Remeis MPICH2 is used for initialisation of MPI tasks which is well supported within Slurm. The process manager is called **pmi2** and is set as default for srun. If an older MPI process manager is needed, for example for older MPI applications used in **torque**, it can be set with | ||
| − | + | <pre> | |
#SBATCH --mpi= | #SBATCH --mpi= | ||
| − | + | </pre> | |
in the submission script. | in the submission script. | ||
| − | + | <pre> | |
srun --mpi=list | srun --mpi=list | ||
| − | + | </pre> | |
provides a list of supported MPI process managers. | provides a list of supported MPI process managers. | ||
| Line 230: | Line 230: | ||
Keep in mind that everything in the compiled programs/scripts which is not an MPI compatible function is executed on each node on its own. For example in ISIS with ''-n 20'': | Keep in mind that everything in the compiled programs/scripts which is not an MPI compatible function is executed on each node on its own. For example in ISIS with ''-n 20'': | ||
| − | + | <pre> | |
fit_counts; | fit_counts; | ||
| − | + | </pre> | |
would fit the defined function to the dataset 20 times at once. That's not very helpful so think about which tasks should be performed in the actual MPI process. Special care has to be taken if something has to be saved as a file. Consider: | would fit the defined function to the dataset 20 times at once. That's not very helpful so think about which tasks should be performed in the actual MPI process. Special care has to be taken if something has to be saved as a file. Consider: | ||
| − | + | <pre> | |
save_par("test.par"); | save_par("test.par"); | ||
| − | + | </pre> | |
with ''-n 20''. This would save the current fit parameters to **test.par** in the working directory 20 times at exactly the same time. This might be helpful if the file is needed on the scratch disk of each node, but doing this on for example ''/userdata'' can cause serious trouble. The function ''mpi_master_only'' can be used to perform a user defined task in an MPI job only once. Best way is to only submit an MPI job to Slurm which only contains actual MPI functions. If some models in ISIS are used which output something to stdout or stderr while loading these messages are also generated 20 times since it's loaded in each process individually. | with ''-n 20''. This would save the current fit parameters to **test.par** in the working directory 20 times at exactly the same time. This might be helpful if the file is needed on the scratch disk of each node, but doing this on for example ''/userdata'' can cause serious trouble. The function ''mpi_master_only'' can be used to perform a user defined task in an MPI job only once. Best way is to only submit an MPI job to Slurm which only contains actual MPI functions. If some models in ISIS are used which output something to stdout or stderr while loading these messages are also generated 20 times since it's loaded in each process individually. | ||
==== Usage ==== | ==== Usage ==== | ||
If the job is a valid MPI process then the submission works exactly like for any other job: | If the job is a valid MPI process then the submission works exactly like for any other job: | ||
| − | + | <pre> | |
#!/bin/bash | #!/bin/bash | ||
#SBATCH --job-name my_first_mpi_job | #SBATCH --job-name my_first_mpi_job | ||
| Line 248: | Line 248: | ||
cd /my/working/dir | cd /my/working/dir | ||
srun /usr/bin/nice -n +15 ./my_mpi_script | srun /usr/bin/nice -n +15 ./my_mpi_script | ||
| − | + | </pre> | |
It might be necessary to set a higher memory usage than for the according non MPI job since some applications try to limit the network traffic by just copying the required data to each node in the first place. | It might be necessary to set a higher memory usage than for the according non MPI job since some applications try to limit the network traffic by just copying the required data to each node in the first place. | ||
| Line 256: | Line 256: | ||
Of course it's also possible to run the MPI process directly from the commandline. As an example let's have a look at the calculation of pi with the MPI program ''cpi''. The program comes with the source code of MPICH2 and is compiled in the ''check'' rule. It's located in | Of course it's also possible to run the MPI process directly from the commandline. As an example let's have a look at the calculation of pi with the MPI program ''cpi''. The program comes with the source code of MPICH2 and is compiled in the ''check'' rule. It's located in | ||
| − | + | <pre> | |
/data/system/software/mpich/mpich-3.2/examples | /data/system/software/mpich/mpich-3.2/examples | ||
| − | + | </pre> | |
To run the calculation in 10 parallel processes directly from the commandline use: | To run the calculation in 10 parallel processes directly from the commandline use: | ||
| − | + | <pre> | |
[1:11]weber@lynx:/data/system/software/mpich/mpich-3.2/examples> srun -n 10 ./cpi | [1:11]weber@lynx:/data/system/software/mpich/mpich-3.2/examples> srun -n 10 ./cpi | ||
Process 0 of 10 is on aquarius | Process 0 of 10 is on aquarius | ||
| Line 274: | Line 274: | ||
pi is approximately 3.1415926544231256, Error is 0.0000000008333325 | pi is approximately 3.1415926544231256, Error is 0.0000000008333325 | ||
wall clock time = 0.010601 | wall clock time = 0.010601 | ||
| − | + | </pre> | |
As we can see Slurm launched 10 processes distributed to aquarius, ara, asterion, aranea and cancer. Keep in mind that running MPI interactively doesn't really make sense. The best way to go is to write a submission script like explained above and let Slurm handle the initialisation. | As we can see Slurm launched 10 processes distributed to aquarius, ara, asterion, aranea and cancer. Keep in mind that running MPI interactively doesn't really make sense. The best way to go is to write a submission script like explained above and let Slurm handle the initialisation. | ||
Revision as of 15:28, 11 April 2018
Copying in progess (Philipp)
SLURM
About
In order to spread the workload of scientific computations on our compute nodes the resource manager SLURM is used.
From [SLURM website]:
Slurm is an open source, fault-tolerant, and highly scalable cluster management and job scheduling system for large and small Linux clusters. Slurm requires no kernel modifications for its operation and is relatively self-contained. As a cluster workload manager, Slurm has three key functions. First, it allocates exclusive and/or non-exclusive access to resources (compute nodes) to users for some duration of time so they can perform work. Second, it provides a framework for starting, executing, and monitoring work (normally a parallel job) on the set of allocated nodes. Finally, it arbitrates contention for resources by managing a queue of pending work.
From [SLURM website]
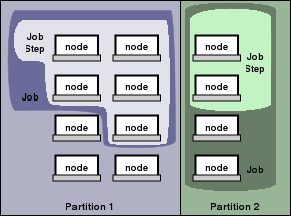
SLURM manages the cluster in partitions, which are a set of compute nodes. Note, that partitions may overlap, e.g. one compute node may be in two or more partitions. A node is a physical computer which provides consumable resources: CPUs and Memory. A CPU does not necessarily have to be a physical processor but is more like a virtual CPU to run one single task on. A dual core with hyper threading technology, for instance, would show up as a node with 4 CPUs consisting of two cores with the capability of running two threads on each core. Physical memory is defined in MB.
The following partitions exist in the current setup:
* remeis: default partition, all machines, timelimit: 14days * erosita: only available for selected people involved in the project, timelimit: infinite * power: only the newest machines and servers, higher priority than 'remeis' (e.g. if you submit via power you will get the power machines as soon as possible and not compete with 'remeis' jobs but only other jobs submitted to 'power'), timelimit: 1day * messier: only messier cluster, also higher priority than 'remeis', timelimit: 7 days * debug: very high priority partition for software development, timelimit: 1h
Quick users tutorial
This tutorial will give you a quick overview over the most important commands. The [SLURM website] provides more detailed information.
Get cluster status
In order to get an overview of the cluster, type
sinfo
This command offers a variety of options how to format the output. In order to get a detailed output while focusing on the nodes rather than the partitions, type
sinfo -N -l
An overview over the available partitions can be shown with
scontrol show partition
an the current queued and running jobs can be displayed using
squeue
Executing a job in real-time
In order to allocate for instance 1 CPUs and 100MB of memory for real-time work, type
salloc --mem-per-cpu=100 -n1 bash
Your bash is now connected to the compute nodes. In order to execute a script use the srun-command
srun my_script.sh
Each srun-command you execute now is interpreted as a job step. The currently running job step of submitted jobs can be displayed using
squeue -s
You can also start a job by simply use the srun command and specify your requirements. In the following case, srun will allocate 100MB of memory and 1 CPU(s) for 1 task, only for the duration of execution.
srun --mem-per-cpu=100 -n1 my_script.sh
If resources are available your job will start immediately.
Submitting a job for later execution
The most convenient way is to submit a job script for later execution. The top part of the script contains scheduling information for SLURM, the more information you provide here, the better.
First of all, a job name is specified, followed by a maximum time. If your job exceeds this time, it will be killed. However, do not overestimate too much because short jobs might start earlier. Acceptable time formats include "minutes", "minutes:seconds", "hours:minutes:seconds", "days-hours", "days-hours:minutes" and "days-hours:minutes:seconds". The output file is set to be test-job(jobID).out and the partition to run the job on is "remeis". The sbatch-script itself will not initiate any job but only allocate the resources. The ntasks and mem-per-cpu options advise the SLURM controller that job steps run within the allocation will launch a maximum of number tasks and to provide for sufficient resources.
The srun commands in the job script launch the job steps. The example below thus consists of two job steps. Each of the srun commands may have own requirements concerning memory any may also spawn less tasks than given in the header of the script file. However, the values in the header may never be exceeded!
#!/bin/bash #SBATCH --job-name my_first_job #SBATCH --time 05:00 #SBATCH --output test-job_%A_%a.out #SBATCH --error test-job_%A_%a.err #SBATCH --partition=remeis #SBATCH --ntasks=4 #SBATCH --mem-per-cpu=100 srun -l my_script1.sh srun -l my_script2.sh
The -l parameter of srun will print the task number in front of a line of stdout/err. You can submit this script by saving it in a file, e.g. my_first_job.slurm, and sumitting it using
cepheus:~> sbatch my_first_job.slurm Submitted batch job 144
You can check the estimated starting time of your job using
squeue --start
Submitting a job array
In order to submit a array of jobs with the same requirements you have to modify your script file. The script above is going to spawn 4 jobs each consisting of one srun command. Note the presence of the new environment variable ${SLURM_ARRAY_TASK_ID} which might be useful for your work. In this example we start an isis-script with different input values. You can also simply use different scripts.
#!/bin/bash
#SBATCH --partition remeis
#SBATCH --job-name important_job
#SBATCH --ntasks=1
#SBATCH --time 00:05:00
#SBATCH --output /home/dauser/tmp/jobscript_beta.%A_%a.out
#SBATCH --error /home/dauser/tmp/jobscript_beta.%A_%a.err
#SBATCH --array 0-3
cd /home/user/script/
COMMAND[0]="./sim_script.sl 0.00"
COMMAND[1]="./sim_script.sl 0.10"
COMMAND[2]="./sim_script.sl 0.20"
COMMAND[3]="./sim_script.sl 0.30"
srun /usr/bin/nice -n +19 ${COMMAND[$SLURM_ARRAY_TASK_ID]}
As above, this code might be saved in a file, for example job.slurm can be executed using
sbatch job.slurm
If you need a specific machine to run your job on, you can use
#SBATCH --nodelist=leo,draco
If you have a job with high I/O and/or traffic on the network you can limit the number of jobs running simultaneously (to 2 in this example) by
#SBATCH --array 0-3%2
SLURM will only allocate resources on the given nodes. However, if all nodes in 'nodelist' cannot fulfill the job requirements, SLURM will also allocate other machines.
If you would like to cancel jobs 1, 2 and 3 from job array 20 use
scancel 20_[1-3]
If you want to cancel the whole array, scancel works as usual
scancel 20
Note, there is also the option to modify requirements of single jobs later using scontrol update job=101_1 ....
If you have jobs which are dependent on the result of others or if you want a more detailed description concerning job arrays you can find it in the official SLURM manual: [[1]]
Submitting a job array where each command needs to change into a different directory
In order to allow each command of the job array to change into an individual directory (as opposed to all into the same directory as above), modify the script as follows:
#!/bin/bash
#SBATCH --partition remeis
#SBATCH --job-name important_job
#SBATCH --ntasks=1
#SBATCH --time 00:05:00
#SBATCH --output /home/dauser/tmp/jobscript_beta.%A_%a.out
#SBATCH --error /home/dauser/tmp/jobscript_beta.%A_%a.err
#SBATCH --array 0-3
DIR[0]="/home/user/dir1"
DIR[1]="/home/user/dir2"
DIR[2]="/userdata/user/dir3"
DIR[3]="/userdata/user/dir4"
cd ${DIR[$SLURM_ARRAY_TASK_ID]}
COMMAND[0]="./sim_script.sl 0.00"
COMMAND[1]="./sim_script.sl 0.10"
COMMAND[2]="./sim_script.sl 0.20"
COMMAND[3]="./sim_script.sl 0.30"
srun /usr/bin/nice -n +19 ${COMMAND[$SLURM_ARRAY_TASK_ID]}
This also works with paths relative to the directory where the slurm script was submitted.
Using the erosita partition (serpens)
If you are allowed to use the eRosita Partition, contact a SLURM admin (eg. [[2]]). Once your username is added to the list of privileged users, you just have to add
#SBATCH --partition=erosita #SBATCH --account=erosita
to your jobfiles.
Other useful commands
* sstat Real-time status information of your running jobs
* sattach <jobid.stepid> Attach to stdI/O of one of your running jobs
* scancel [OPTIONS...] [job_id[_array_id][.step_id]] [job_id[_array_id][.step_id]...] Cancel the execution of one of your job arrays/jobs/job steps.
* scontrol Administration tool, you can for example use this to modify the requirements of your jobs. You can for exaple show your jobs show jobs or update the time limit update JobId= TimeLimit=2.
* smap graphically view information about Slurm jobs, partitions, and set configurations parameters.
* sview graphical user interface for those who prefer clicking over typing. X-Server required.
SLURM and MPI
About MPI
MPI (the Message Passing Interface) makes it possible to run parallel processes on CPUs of different hosts. To do so it uses TCP packets to communicate via the normal network connection. Some tasks can profit a lot of using more cores for computation. At Remeis MPICH2 is used for initialisation of MPI tasks which is well supported within Slurm. The process manager is called **pmi2** and is set as default for srun. If an older MPI process manager is needed, for example for older MPI applications used in **torque**, it can be set with
#SBATCH --mpi=
in the submission script.
srun --mpi=list
provides a list of supported MPI process managers.
The implementation of MPI for SLang/ISIS is called **SLMPI**.
Best practice for MPI tasks
The usage of MPI might cause continuously high network traffic especially on the host which holds the master process. Please consider this when deciding which nodes are used for the job. It's a good idea to provide servers (e.g. leo or lupus) with the --nodelist= option one of which is then used to hold the master process since nobody is sitting in front of it and trying to use a browser. Additional nodes are allocated automatically by Slurm if required to fit the --ntasks / -n option.
MPI jobs depend on all allocated nodes to be up and running properly, so I'd like to use this opportunity to remind about shutting down/rebooting PCs on your own without any permission can abort a whole MPI job.
Requirements and Tips
To use MPI obviously the application or function used should support MPI. Examples range from programs written in C using some MPI features and compiled with the mpicc compiler to common ISIS-functions such as mpi_emcee or mpi_fit_pars.
Keep in mind that everything in the compiled programs/scripts which is not an MPI compatible function is executed on each node on its own. For example in ISIS with -n 20:
fit_counts;
would fit the defined function to the dataset 20 times at once. That's not very helpful so think about which tasks should be performed in the actual MPI process. Special care has to be taken if something has to be saved as a file. Consider:
save_par("test.par");
with -n 20. This would save the current fit parameters to **test.par** in the working directory 20 times at exactly the same time. This might be helpful if the file is needed on the scratch disk of each node, but doing this on for example /userdata can cause serious trouble. The function mpi_master_only can be used to perform a user defined task in an MPI job only once. Best way is to only submit an MPI job to Slurm which only contains actual MPI functions. If some models in ISIS are used which output something to stdout or stderr while loading these messages are also generated 20 times since it's loaded in each process individually.
Usage
If the job is a valid MPI process then the submission works exactly like for any other job:
#!/bin/bash #SBATCH --job-name my_first_mpi_job #SBATCH ... #SBATCH --ntasks=20 cd /my/working/dir srun /usr/bin/nice -n +15 ./my_mpi_script
It might be necessary to set a higher memory usage than for the according non MPI job since some applications try to limit the network traffic by just copying the required data to each node in the first place.
Also make sure that if it is necessary to specify the number of child processes in the application itself, set it to the same as with the --ntasks / -n option in the submission. An example would be the num_slaves qualifier in mpi_emcee.
Note that the srun command does not contain mpiexec or mpirun which were used in older versions of MPI to launch the processes. The processes manager pmi2 is built into Slurm and makes it possible that Slurm itself can initialize the network communication with the srun command only.
Of course it's also possible to run the MPI process directly from the commandline. As an example let's have a look at the calculation of pi with the MPI program cpi. The program comes with the source code of MPICH2 and is compiled in the check rule. It's located in
/data/system/software/mpich/mpich-3.2/examples
To run the calculation in 10 parallel processes directly from the commandline use:
[1:11]weber@lynx:/data/system/software/mpich/mpich-3.2/examples> srun -n 10 ./cpi Process 0 of 10 is on aquarius Process 1 of 10 is on ara Process 6 of 10 is on asterion Process 2 of 10 is on ara Process 8 of 10 is on asterion Process 7 of 10 is on asterion Process 3 of 10 is on aranea Process 5 of 10 is on aranea Process 4 of 10 is on aranea Process 9 of 10 is on cancer pi is approximately 3.1415926544231256, Error is 0.0000000008333325 wall clock time = 0.010601
As we can see Slurm launched 10 processes distributed to aquarius, ara, asterion, aranea and cancer. Keep in mind that running MPI interactively doesn't really make sense. The best way to go is to write a submission script like explained above and let Slurm handle the initialisation.