Difference between revisions of "Plotting Model Components (xfig example)"
| Line 11: | Line 11: | ||
=== Loading the data === | === Loading the data === | ||
| − | < | + | <pre> |
load_data("ridge_epoch_4_2.pha"); % load your spectra | load_data("ridge_epoch_4_2.pha"); % load your spectra | ||
| Line 19: | Line 19: | ||
ignore_en(1,20, ); | ignore_en(1,20, ); | ||
| − | </ | + | </pre> |
=== Starting the plot === | === Starting the plot === | ||
| − | < | + | <pre> |
open_plot; % open a plot window... | open_plot; % open a plot window... | ||
multiplot([5,2]); % ...to get the two panels for spectrum and residuals | multiplot([5,2]); % ...to get the two panels for spectrum and residuals | ||
| Line 40: | Line 40: | ||
% plot it will end in .dat | % plot it will end in .dat | ||
| − | </ | + | </pre> |
and now a single component of the model: | and now a single component of the model: | ||
| − | < | + | <pre> |
ff = get_fit_fun(); % save the complete model | ff = get_fit_fun(); % save the complete model | ||
| Line 54: | Line 54: | ||
write_plot("write_data_gauss"); % again, dump the plot into another ASCII file | write_plot("write_data_gauss"); % again, dump the plot into another ASCII file | ||
| − | </ | + | </pre> |
Of course the colors are unimportant for the moment, as you will have to do them again when doing the plot in xfig, but now the example is complete if you don't have the time for xfig and just want a quick way to plot. | Of course the colors are unimportant for the moment, as you will have to do them again when doing the plot in xfig, but now the example is complete if you don't have the time for xfig and just want a quick way to plot. | ||
| Line 60: | Line 60: | ||
=== The second panel === | === The second panel === | ||
| − | < | + | <pre> |
mpane(2); % move to the lower panel | mpane(2); % move to the lower panel | ||
ylin; % for residuals, a linear y scale | ylin; % for residuals, a linear y scale | ||
| Line 72: | Line 72: | ||
write_plot("write_res_allin"); % dump the plot into an ASCII file. for residual plots | write_plot("write_res_allin"); % dump the plot into an ASCII file. for residual plots | ||
% it will end in .res | % it will end in .res | ||
| − | </ | + | </pre> |
Now the residuals you get when the component is missing in the model (in this case: an iron line complex consisting of three gaussian lines): | Now the residuals you get when the component is missing in the model (in this case: an iron line complex consisting of three gaussian lines): | ||
| − | < | + | <pre> |
set_par([5,8,11],0); % set the norm of your component to zero | set_par([5,8,11],0); % set the norm of your component to zero | ||
() = eval_counts; % evaluate the model | () = eval_counts; % evaluate the model | ||
| Line 83: | Line 83: | ||
write_plot("write_res_gauss.dat"); % dump the plot into an ASCII file | write_plot("write_res_gauss.dat"); % dump the plot into an ASCII file | ||
close_plot; % close the plot window when finished | close_plot; % close the plot window when finished | ||
| − | </ | + | </pre> |
Now you have all the files you need to make a nice xfig plot. | Now you have all the files you need to make a nice xfig plot. | ||
| Line 90: | Line 90: | ||
=== Initializing the plot === | === Initializing the plot === | ||
| − | < | + | <pre> |
require("xfig"); % you need this to do the nice plots | require("xfig"); % you need this to do the nice plots | ||
| Line 105: | Line 105: | ||
p[1].ylabel(`$\chi$`); % ylabel of the lower panel | p[1].ylabel(`$\chi$`); % ylabel of the lower panel | ||
p[1].xlabel("Energy [keV]"); % xlabel of the whole plot | p[1].xlabel("Energy [keV]"); % xlabel of the whole plot | ||
| − | </ | + | </pre> |
=== Read and plot the ASCII files: upper panel === | === Read and plot the ASCII files: upper panel === | ||
| − | < | + | <pre> |
variable lo, hi, y, e, m; % five variables for the five columns of the .dat file | variable lo, hi, y, e, m; % five variables for the five columns of the .dat file | ||
(lo, hi, y, e, m) = readcol ("write_data_allin_0.dat",1,2,3,4,5); % read all five columns: bin_lo, bin_hi, value, error, model | (lo, hi, y, e, m) = readcol ("write_data_allin_0.dat",1,2,3,4,5); % read all five columns: bin_lo, bin_hi, value, error, model | ||
| Line 123: | Line 123: | ||
p[0].plot (x,y,e; sym="point"); % a normal plot to get data points and errorbars | p[0].plot (x,y,e; sym="point"); % a normal plot to get data points and errorbars | ||
| − | </ | + | </pre> |
now the model component: | now the model component: | ||
| − | < | + | <pre> |
variable lo, hi, y, e, m; % clear the variables the previous variables | variable lo, hi, y, e, m; % clear the variables the previous variables | ||
(lo, hi, y, e, m) = readcol ("write_data_gauss_0.dat",1,2,3,4,5); % read the ASCII file with the component | (lo, hi, y, e, m) = readcol ("write_data_gauss_0.dat",1,2,3,4,5); % read the ASCII file with the component | ||
| Line 133: | Line 133: | ||
p[0].hplot (h,m; color="green4"); % plot a histogram of the model component | p[0].hplot (h,m; color="green4"); % plot a histogram of the model component | ||
| − | </ | + | </pre> |
=== Read and plot the ASCII files: lower panel === | === Read and plot the ASCII files: lower panel === | ||
| − | < | + | <pre> |
variable lo, hi, y, el, eu; % five variables for the columns of the .res file | variable lo, hi, y, el, eu; % five variables for the columns of the .res file | ||
(lo, hi, y, el, eu) = readcol ("write_res_allin_0.res",1,2,3,4,5); % read all five columns: bin_lo, bin_hi, value, err_lo, err_hi | (lo, hi, y, el, eu) = readcol ("write_res_allin_0.res",1,2,3,4,5); % read all five columns: bin_lo, bin_hi, value, err_lo, err_hi | ||
| Line 147: | Line 147: | ||
% optional: add the errorbars, if you did the plot with an | % optional: add the errorbars, if you did the plot with an | ||
% individually sized last bin (i.e., added the last bin_hi, see above) | % individually sized last bin (i.e., added the last bin_hi, see above) | ||
| − | </ | + | </pre> |
now the residuals without the component: | now the residuals without the component: | ||
| − | < | + | <pre> |
variable lo, hi, y, el, eu; % clear the variables | variable lo, hi, y, el, eu; % clear the variables | ||
(lo, hi, y, el, eu) = readcol ("write_res_gauss_0.res",1,2,3,4,5); % read the .res file | (lo, hi, y, el, eu) = readcol ("write_res_gauss_0.res",1,2,3,4,5); % read the .res file | ||
| Line 161: | Line 161: | ||
p[1].plot (x,y, ,{y-el,eu-y}; sym="point", color="green4", size=-1, eb_factor=0); | p[1].plot (x,y, ,{y-el,eu-y}; sym="point", color="green4", size=-1, eb_factor=0); | ||
% optional: add the errorbars | % optional: add the errorbars | ||
| − | </ | + | </pre> |
=== Render the plot === | === Render the plot === | ||
| − | < | + | <pre> |
xfig_multiplot (p).render ("plot_components.png"); | xfig_multiplot (p).render ("plot_components.png"); | ||
| − | </ | + | </pre> |
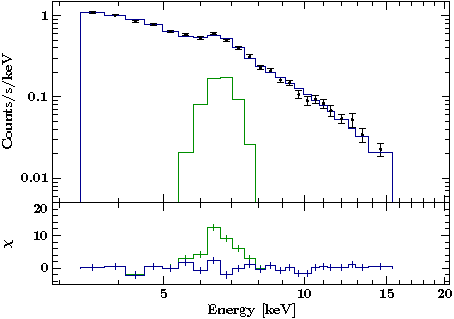
===== The Result ===== | ===== The Result ===== | ||
Revision as of 18:01, 13 April 2018
Plotting Model Components
Even in ISIS, plotting individual model components is somewhat tricky. Once upon a time, there was a useful description to be found [[1]] on pp. 39-43. Unfortunately, since then, the fancy plot commands changed but not the presentation. I will try here to give you a short overview of how to do this at the moment.
The plot in ISIS
Loading the data
load_data("ridge_epoch_4_2.pha"); % load your spectra
flux_corr(1); % the usual setup things
group(1; min_sn=5, min_chan=1, bounds=3, unit="keV");
ignore_en(1, ,3);
ignore_en(1,20, );
Starting the plot
open_plot; % open a plot window...
multiplot([5,2]); % ...to get the two panels for spectrum and residuals
mpane(1); % start plotting in the upper panel
xlog; ylog; % if that's the usual way you want to have your spectra
load_par("2011-03-09_15:00:21_best.par"); % load the best-fit parameters for the whole model
() = eval_counts; % evaluate the model
plot_data(1; dsym=-4, dcol=1, decol=1, mcol=15, xrange={2.9,20.5}, yrange={0.005,1.5}, no_reset=1);
% the command to plot your spectrum. note that the
% "no_reset" command now has to be used as a qualifier,
% different from Mike's presentation. also, the commands
% for setting the ranges have changed (from "xrng" to "xrange").
write_plot("write_data_allin"); % dump the spectrum and model into an ASCII file. for a data
% plot it will end in .dat
and now a single component of the model:
ff = get_fit_fun(); % save the complete model
fit_fun("gauss(1)"); % now only the component you are interested in
% **ATTENTION**: next time, reload the best-fit parameters before
% adding another model component: "fit_fun(ff)"!!!
() = eval_counts; % evaluate the model (= the single component)
plot_data(1; dsym=-4, dcol=1, decol=1, mcol=4, oplt=1, no_reset=1); % plot the component in a different color
write_plot("write_data_gauss"); % again, dump the plot into another ASCII file
Of course the colors are unimportant for the moment, as you will have to do them again when doing the plot in xfig, but now the example is complete if you don't have the time for xfig and just want a quick way to plot.
The second panel
mpane(2); % move to the lower panel
ylin; % for residuals, a linear y scale
load_par("2011-03-09_15:00:21_best.par"); % again, reload the best-fit parameters
() = eval_counts; % evaluate the model
plot_residuals(1; res=1, rsym=0, rcol=15, recol=1, xrange={2.9,20.5}, yrange={-3,20}, no_reset=1);
% plot only the residuals. qualifier are the
% same as for "plot_data"
write_plot("write_res_allin"); % dump the plot into an ASCII file. for residual plots
% it will end in .res
Now the residuals you get when the component is missing in the model (in this case: an iron line complex consisting of three gaussian lines):
set_par([5,8,11],0); % set the norm of your component to zero
() = eval_counts; % evaluate the model
plot_residuals(1; res=1, rsym=0, rcol=4, recol=1, oplt=1, no_reset=1);
% plot the residuals
write_plot("write_res_gauss.dat"); % dump the plot into an ASCII file
close_plot; % close the plot window when finished
Now you have all the files you need to make a nice xfig plot.
Reading and plotting the data with xfig
Initializing the plot
require("xfig"); % you need this to do the nice plots
variable p = Struct_Type[2]; % this way, you can do a multiplot
p[0] = xfig_plot_new(12.6,6.4); % the size of the upper panel
p[1] = xfig_plot_new(12.6,2.6); % size of the lower panel. proportions are the same as in ISIS plots.
p[0].world (2.9,20.5,0.005,1.5; xlog=1, ylog=1); % ranges and scales of the upper panel
p[0].x1axis(;ticlabels=0); % no ticlabels between the two panels
p[0].y2axis(;ticlabels=0); % no ticlabels at the right side of the plot
p[0].ylabel("Counts/s/keV"); % ylabel of the upper panel
p[1].world (2.9,20.5,-5,41; xlog=1); % ranges and scales of the lower panel
p[1].ylabel(`$\chi$`); % ylabel of the lower panel
p[1].xlabel("Energy [keV]"); % xlabel of the whole plot
Read and plot the ASCII files: upper panel
variable lo, hi, y, e, m; % five variables for the five columns of the .dat file
(lo, hi, y, e, m) = readcol ("write_data_allin_0.dat",1,2,3,4,5); % read all five columns: bin_lo, bin_hi, value, error, model
variable h = [lo, hi[-1]]; % add the upper limit of the last bin to the lower limits of the bins
variable x = (lo+hi)/2; % an array with the central points of each bin
p[0].hplot (h,m; color="blue4"); % make a histogram plot of the model
% see "p[0].hplot(;help)" for help:
% if length(h)=length(m)+1, the last element is taken as the upper
% limit of the last bin. if both arguments have the same length,
% the last bin will have the same size as the one before.
% **ATTENTION** at the moment, hplot can only plot a histogram with
% errorbars when all arrays have the same length!!!
p[0].plot (x,y,e; sym="point"); % a normal plot to get data points and errorbars
now the model component:
variable lo, hi, y, e, m; % clear the variables the previous variables
(lo, hi, y, e, m) = readcol ("write_data_gauss_0.dat",1,2,3,4,5); % read the ASCII file with the component
variable h = [lo, hi[-1]]; % add the last bin_hi for histogram plotting
p[0].hplot (h,m; color="green4"); % plot a histogram of the model component
Read and plot the ASCII files: lower panel
variable lo, hi, y, el, eu; % five variables for the columns of the .res file
(lo, hi, y, el, eu) = readcol ("write_res_allin_0.res",1,2,3,4,5); % read all five columns: bin_lo, bin_hi, value, err_lo, err_hi
variable h = [lo, hi[-1]]; % add the last bin_hi for histogram plotting
variable x = (lo+hi)/2; % the center of each bin
p[1].hplot (h,y; color="blue4"); % plot the residuals histogram of the whole model
p[1].plot (x,y, ,{y-el,eu-y}; sym="point", color="blue4", size=-1, eb_factor=0);
% optional: add the errorbars, if you did the plot with an
% individually sized last bin (i.e., added the last bin_hi, see above)
now the residuals without the component:
variable lo, hi, y, el, eu; % clear the variables
(lo, hi, y, el, eu) = readcol ("write_res_gauss_0.res",1,2,3,4,5); % read the .res file
variable h = [lo, hi[-1]]; % add the last bin_hi for histogram plotting
variable x = (lo+hi)/2; % center of each bin
p[1].hplot (h,y; color="green4"); % plot the residuals histogram without the component\
p[1].plot (x,y, ,{y-el,eu-y}; sym="point", color="green4", size=-1, eb_factor=0);
% optional: add the errorbars
Render the plot
xfig_multiplot (p).render ("plot_components.png");
The Result
To remove the annoying vertical lines at the first and last bin of the histogram, you can use the (undocumented) qualifiers y_last and y_first of hplot. If you set them to the value of the first or the last bin respectively, the vertical lines vanish:
p[0].hplot (h,m; color="blue4", y_first=m[0], y_last=m[-1]);