Integral
INTEGRAL Scripts
The Remeis INTEGRAL scripts can be found under
/software/Science/satscripts/intscripts/
on Remeis machines. If you have any questions on how to use them, read the following text and talk to Ingo or Philipp.
General remarks on Integral data extraction at Remeis:
Initially data is extracted in parallel on a ScWs basis and in a second step combined to the final products. This allows for the usage of SLURM and leads to significantly faster extraction compared to a combined extraction on a single machine. The initial step is launch by the scripts master_sbs. The script can generate spectra and images for ISGRI and spectra for JEMX. No PICsIT, SPI or OMC analysis is run at Remeis. The resulting spectra are then combined using spe_pick while the ISGRI/IBIS images are merged using INT_mosa_ibis. As the spectral extraction requires a catalog containing the sources for which the spectral extraction should be run, it is advised to first generate images & a mosaic to determine, which sources are bright and detected in the field of view. Generally it is a good idea, if you have not already to have a look at the OSA cookbook, e.g., to check how the paths in the data object locator (DOL) files is supposed to look like or how to prepare a suitable catalog for spectral extraction: https://www.isdc.unige.ch/integral/analysis
Before any OSA analysis can be run the corresponding module has to be load with:
ml integral-osa
Before running any data extraction you need to set up a few soft links:
ln -sf /home/X-ray/INTEGRAL/archive/aux ln -sf /home/X-ray/INTEGRAL/archive/ic ln -sf /home/X-ray/INTEGRAL/archive/idx ln -sf /home/X-ray/INTEGRAL/archive/scw
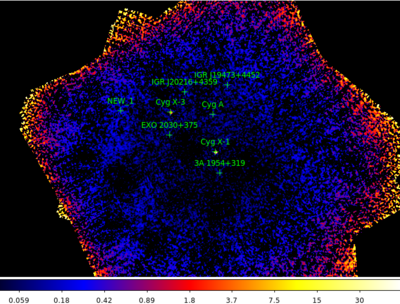
Typical IBIS image/mosaic
- 1) Find ScWs that cover your source, e.g., pointing is < 10 degree away from source
objscw --scwlist "Cyg X-1" --maxdist=10 > scws_all.txt
If you want to use scws_all.txt as input for hte next step remove the header of this file, i.e, the first 6 lines. Importantly you will probalby want to select only a subset of the available observations and therefore perform some selection on the full list of science windows. As a simple example one might image to only use the latest 100 ScWs:
tail -n 100 scws_all.txt > scws_cut.txt
- 2) Start the ScW based analysis
master_sbs --job=IMA --scwlist=scws_all.txt --energies="30,50,200" --verbose --clobber=yes --path=$PWD
- 3) Generate the required input file for spe_pick
txt2idx element="scws_dols.txt" index="cygx1"
- 4) Run INT_mosa_ibis to generate the a combine mosaic
INT_mosa_ibis --obsgrp=$PWD --dol=scws_dols.txt --destin=$PWD/obs/mosa_cygx1
- 5) Check the images for artifacts and sort out affected ScWs through trial-and-error
ds9 isgri_mosa_ima.fits[2] -region found.reg -cmap b -scale sqrt -scale limits 0 60 -zoom 2
The resulting image should look something like this:
Details about the detected sources, such as there flux and detection significance can be found in the file isgri_mosa_res.fits.
Typical IBIS spectral analysis
- 1) Again, find ScWs that cover your source and are of interest to you
- 2) Start the ScW based analysis
master_sbs --job=SPE --scwlist=scws_cut.txt --source="Cyg X-1" --verbose --clobber=yes --path=$PWD --catalog=specat.fits
- 3) Generate the required input file for spe_pick. Check that that all objects in the DOL file exist.
txt2idx element="scws_dols.txt" index="cygx1"
- 4) Run spe_pick to generate the final spectra
spe_pick group="cygx1.fits" source="Cyg X-1" rootname="cygX1_all" instrument="ISGRI"
This will result in background subtracted spectra for the given source. Remember that the source name has exactly the naming in the INTEGRAL reference catalog.
Typical IBIS light-curve generation
- 1) Quite similar to the spectral extraction. First find ScWs that cover your source, or reuse the previous ScW list
- 2) Start the ScW based analysis, here for 100s time binning
master_sbs --job=LC --scwlist=scws_cut.txt --source="Cyg X-1" --verbose --clobber=yes --path=$PWD --catalog=specat.fits --energies=30,120 --deltat=100
This generates a separate light-curve for each ScW at, e.g., obs/250300170010/scw/250300170010.001/250300170010_Cyg_X-1_30-120.lc
- 3) These can then be merged together e.g. using fmerge or ISIS.
ls obs/*/scw/*.001/*_Cyg_X-1_30-120.lc >> lcs.txt fmerge @lcs.txt Cyg_X1_ibis_lc.fits -
Typical IBIS PIF light-curve generation
The above light-curves are only reliable down to a time-resolution of 1.0-0.1s. For higher timing resolution the deconvolution becomes unstable and one has to generate light-curves from the event-lists and pixel-illumination fraction (PIF).
- 1) First event lists are generated with the respective Remeis Scripts
eventlist --path=$PWD --source="Cyg X-1" --scw=$scw --catalog=$cat
This generates a event-list under "obs/266100220010/scw/266100220010.001/events.fits". For each source in the catalog and the FOV this file includes a column indicating whether a source is illuminated by the respective source.
- 2) Filter the eventfile e.g. with an ftools or an ISIS.
fcopy "events.fits[2][PIF_1==1]" cyg_pif1.fits
- 3) Light-curves can then be generated e.g. with ISIS (lc_from_events)
variable lc = lc_from_events(struct{time=evts.time[n], pi=evts.energy[n]};dt=dt,
gti = fits_read_table(gtiname + "[14]"), minfracexp=.5);
Typical JEMX spectral analysis
Until revolution 976 (Oct. 10,2010) only one JEM-X unit was operating at any give time. During that time one should therefore run the analysis for both detectors.
- 1) Find ScWs that cover your source. As JEMX has a smaller field of view, its max distance to the source should also be reduced to ~5 degree
objscw --scwlist "Cyg X-1" --maxdist=5 > scws_jmx.txt
And again, you will probably further select in which ScWs you are interested.
- 2) Generate a RMF with the required binning, e.g., 32 bins
j_rebin_rmf binlist=STD_032
The RMF does not change between observations and might be reused as long as binning requirements do not change. (Depending on your fitting software the resulting RMF might give you troubles and needs to be rewritten.)
- 3) Start the ScW based analysis with 32=2^5 logarithmic energy bins
master_sbs --path=$PWD --scwlist=scws_jemx_cut.txt --job=SPE --source="Cyg X-1" --jemx=1 --clobber=yes --catalog=catalog_jemx.fits --response=jemx_rebinned_rmf.fits --nChanBins=-5
- 4) Generate the required input file for spe_pick
txt2idx element="scws_jmx_dols.txt" index="cygx1"
- 5) Run spe_pick to generate the final spectra.
spe_pick group=cygx1.fits source="Cyg X-1" rootname="cygx1" instrument="JMX1"
JEMX images
To generate JEMX images there are no dedicated scripts at Remeis, and you have to fall back to the OSA scripts, e.g.:
og_create idxSwg=scw.lst ogid=oph2 baseDir="./" instrument=JMX1 jemx_science_analysis startLevel="IMA2" endLevel="IMA2" jemxNum=1 COR_gainModel=2
On catalogs
Adapted from the documentation: While for generally image generation a user generated catalog is not required, if supplied, it needs to keeps the same format of the general reference catalog from which it was built, i.e. it has the extension of the type ”GNRL-REFR-CAT”. This is the correct extension to be used as input for the Imaging step. In Section 7.2 of the OSA manual, you are been shown how to build a catalog for the lightcurve (LCR) and spectral (SPE) step. In that case the catalog, you obtained, specat.fits, was of the type of ”ISGR-SRCLRES” which is the correct format for the LCR and SPE steps. The two formats cannot be switched, meaning that you cannot use a ”GNRL-REFR-CAT” type catalog for SPE and LCR or an ”ISGR-SRCL-RES” one for IMA.
In practice one prepares the catalog used for spectral extraction from the results of the IMA step, taking only into account actually detected sources, e.g. by running the following on the results of INT_mosa_ibis:
fcopy "isgri_srcl_res.fits[1][DETSIG>10]" specat.fits
Time selection
User generated GTI files can be used to perform time or phase resolved spectral analysis. This is done through the --gtiname & --gtiformat keywords. Consult the master_sbs help for more details. To generate GTI file it is recommended to make use of the OSA script gti_user, e.g., as follows:
gti_user tstart=8170.5 tstop=8171.8 unit=sec begin=0.0 end=50 step=150 repeat=653 gti=user_gti.fits clobber=yes
Or for a to calculate a reverse bary-corrected GTI file. Not that this requires the coordinated and the DOL file of a ScW or Observation Group.
gti_user tstart=8393.16 unit=sec begin=0.0 end=50 step=25 repeat=600 gti=user_gti.fits clobber=yes bary=y group=og_ibis.fits+1
Also note that the light-curves generated by imalc_sbs through master_sbs are already barycenter corrected.
More details can be found in the introduction to INTEGRAL analysis:
https://www.isdc.unige.ch/integral/download/osa/doc/11.2/osa_um_intro.pdf
Quick Look Analysis
NRT data can also be analysed at Remeis with the usual scripts, require, however, an additional --nrt parameter in most scripts. Also have a look at http://isdc.unige.ch/heavens/
Documentation
Useful to know: http://www.isdc.unige.ch/integral/download/osa/doc/10.0/osa_um_ibis/node27.html And for the cook-book and general documentation. Reading the IBIS Analysis User Manual is recommend: https://www.isdc.unige.ch/integral/analysis Each INTEGTAL script also has a help
master_sbs --help cat $ISDC_ENV/help/ibis_science_analysis.txt
Abbreviations
http://www.isdc.unige.ch/integral/support/glossary
- DAL The Data Access Layer (DAL) is a software library used to manipulate (create, write, read and manage) scientific data.
- DOL The Data Object Location (DOL) specifies the location of a DAL object, both by the FITS file name and its extension. For instance, /isdc/dev/xxx.fits[2] is a DOL, while xxx.fits is simply the name of a FITS file.
Known Issues
- In general: check first http://www.isdc.unige.ch/integral/support/faq for the error code you get
- The resulting spectra will not be strictly OGIP compliant, which e.g. ISIS might care about, but can be ignored.
Errors mosaicing using custom DOLs
If you want to create a mosaic using DOL (Data Object Location) files, sometimes the building interrupts with errors like a Fortran Runtime error (http://www.isdc.unige.ch/integral/support/faq.cgi?SOFT-051) or errors like:
Using and copying first SCW >>/scratch1/beuchert/integral/scw_extr/obs/125400400010/scw/125400400010.001/swg_ibis.fits << Log_0 : 100 elements opened. Log_0 : 100 elements attached. Error_1: Can not update the index.
or
Error_1: Can not close the element
or
Error_2: DAL_READ_ERROR : error reading from FITS file
etc.
- Somehow corrupted scws? Collect sets of two scws and create Mosaics based on those. Like that it is easy to enclose corrupted scws.
- An error -2004 (DAL_FILE_NOT_ACCESSIBLE) often results because one of your groups is corrupted. Before restarting the analysis, make a ``dal_clean, which should be able to do an automatic repair. If it does not work try a ``dal_verify to see what is corrupted. Or you can try to open your group (or child group) with fv and look for a file pointed by the group that does not exists. Most of the time removing the line with this wrong file in the group table is fixing your problem.
The latter: first create an observation group out of your dol-list of scws. Creating observation groups is also the canonical way of extracting INTEGRAL/IBIS data. Before doing that, the soft links as noted above have to be set in the directory. Then:
cd path/to/dir/with/softlinks/ og_create idxSwg=dol.txt ogid=isgri_gc baseDir="./" instrument=IBIS ls obs/isgri_gc/
Repair the observation group and included indices/links with
dal_clean inDOL="obs/isgri_gc/og_ibis.fits" chatty="4" checkExt="1"
Now INT_mosa_ibis should work again on the scws in dol.txt
How to create mosaics (outdated)
First make a list of scws you want to add to a mosaic, i.e., a "dol-file" dol.txt:
/scratch1/beuchert/integral/scw_extr/obs/066200340010/scw/066200340010.001/swg_ibis.fits /scratch1/beuchert/integral/scw_extr/obs/066200350010/scw/066200350010.001/swg_ibis.fits /scratch1/beuchert/integral/scw_extr/obs/066200360010/scw/066200360010.001/swg_ibis.fits
Then prepare the directory where the mosaic shall be built in, i.e., set the env. variables to allow INT_mosa_ibis to find indices etc.:
cd /some/path/ ln -s /virgo/b/INTEGRAL/aux/ ln -s /virgo/b/INTEGRAL/idx/ ln -s /virgo/b/INTEGRAL/ic/ ln -s /virgo/b/INTEGRAL/scw/ ln -s /virgo/b/INTEGRAL/cat/ mkdir /some/path/obs/ INT_mosa_ibis --obsgrp=/path/to/all/extracted/scws/ --destin=/path/to/the/prepared/directory/obs/mosa_og/ --dol="dol.txt"
Non-OSA INTEGRAL extraction
There are a few alternatives by now to using OSA+Remeis scripts:
- To run IBIS and JEMX analysis online there is MMODA at https://www.astro.unige.ch/mmoda/help/mmoda/integral-mission
- If you are interested in SPI data you can do this with the OSA installed here at Remeis, there are however much more reliable and stable pipelines available in form of either SPI-DAI or the SPI-pipeline at MPE organized by Tomas Stiegert.